Stan is where we onboard teammates to Bayesian workflows. A logistic regression is the gentlest way to see how Stan’s modeling language, inference engine, and diagnostics line up, so this post walks through the full pipeline end-to-end.
Logistic regression in equations
For observation $n = 1, \ldots, N$ with predictors $x_n \in \mathbb{R}^K$ and binary outcome $y_n \in {0, 1}$, the logistic regression model is
$$ \begin{aligned} y_n &\sim \operatorname{Bernoulli}(\theta_n), \ \theta_n &= \operatorname{logit}^{-1}(\eta_n), \ \eta_n &= \alpha + x_n^\top \beta, \end{aligned} $$
where $\alpha$ is the intercept, $\beta \in \mathbb{R}^K$ are slope coefficients, and $\operatorname{logit}^{-1}(z) = \frac{1}{1 + e^{-z}}$. In vector form, with design matrix $X \in \mathbb{R}^{N \times K}$, the linear predictor is $\eta = \alpha \mathbf{1}_N + X \beta$. Here $\mathbf{1}_N$ represents an $N$-dimensional column vector of ones, so $\alpha \mathbf{1}_N$ is simply the intercept repeated for every observation; adding it to $X \beta$ shifts each linear predictor by the same baseline amount. The Stan program below encodes this model along with weakly informative priors on $\alpha$ and $\beta$.
Why Stan for discrete outcomes
- Express the likelihood and priors directly in Stan’s modeling language without wrestling with black-box abstractions.
- Leverage Hamiltonian Monte Carlo (HMC) to explore posteriors that simple Metropolis samplers struggle with.
- Inspect diagnostics (divergences, effective sample sizes, R-hat) that ship by default with every fit.
- Reuse the same Stan program across R, Python, or the command-line interface.
1. Scaffold a minimal Stan project
We’ll use cmdstanpy, which gives Python bindings to the CmdStan interface. Once you install CmdStan (cmdstanpy.install_cmdstan()) the wrapper takes care of compilation and sampling.
from cmdstanpy import CmdStanModel
import pandas as pd
import numpy as np
# Toy admissions dataset: GPA + GRE scores predicting acceptance
students = pd.DataFrame(
{
"gpa": [2.5, 3.2, 3.8, 2.9, 3.6, 3.1, 2.7, 3.9, 3.4, 2.8],
"gre": [480, 600, 720, 520, 700, 640, 500, 760, 660, 540],
"admit": [0, 0, 1, 0, 1, 1, 0, 1, 1, 0],
}
)
students["gre_std"] = (students["gre"] - students["gre"].mean()) / students["gre"].std()
students["gpa_std"] = (students["gpa"] - students["gpa"].mean()) / students["gpa"].std()
Standardizing predictors keeps the sampler happy because the coefficients live on comparable scales.
2. Assemble data for Stan
Stan consumes a dictionary of primitives. Split out the design matrix and response, then feed them to the model.
X = students[["gre_std", "gpa_std"]].to_numpy()
stan_data = {
"N": students.shape[0],
"K": X.shape[1],
"X": X,
"y": students["admit"].to_numpy(),
}
3. Write the Stan program
The Stan file defines the generative story: priors on the intercept and weights, a Bernoulli likelihood with logit link, and a generated quantities block for pointwise log-likelihood (useful for LOO or WAIC later).
data {
int<lower=0> N;
int<lower=1> K;
matrix[N, K] X;
array[N] int<lower=0, upper=1> y;
}
parameters {
real alpha;
vector[K] beta;
}
model {
alpha ~ normal(0, 5);
beta ~ normal(0, 2.5);
y ~ bernoulli_logit(alpha + X * beta);
}
generated quantities {
array[N] real log_lik;
array[N] int<lower=0, upper=1> y_hat;
for (n in 1:N) {
real eta = alpha + X[n] * beta;
log_lik[n] = bernoulli_logit_lpmf(y[n] | eta);
y_hat[n] = bernoulli_logit_rng(eta);
}
}
Keep the file as models/logistic_regression.stan (or similar) inside your project.
4. Compile and sample
With CmdStan installed, compilation translates the Stan program into optimized C++. Sampling launches four HMC chains by default.
model = CmdStanModel(stan_file="models/logistic_regression.stan")
fit = model.sample(
data=stan_data,
iter_warmup=1000,
iter_sampling=1000,
chains=4,
seed=20251030,
)
fit.summary()
You’ll see posterior estimates for the intercept (alpha) and standardized coefficients (beta[1] for GRE, beta[2] for GPA), plus diagnostics like R_hat, bulk/tail ess, and divergence counts.
5. Diagnose and interpret
Stan surfaces rich diagnostics—treat them as required reading, not optional.
fit.diagnose()flags divergences or treedepth saturations that mean the sampler struggled; address them with better priors or reparameterization.- Trace plots (
fit.draws()) reassure you that chains mix and explore the posterior rather than sticking to a single mode.
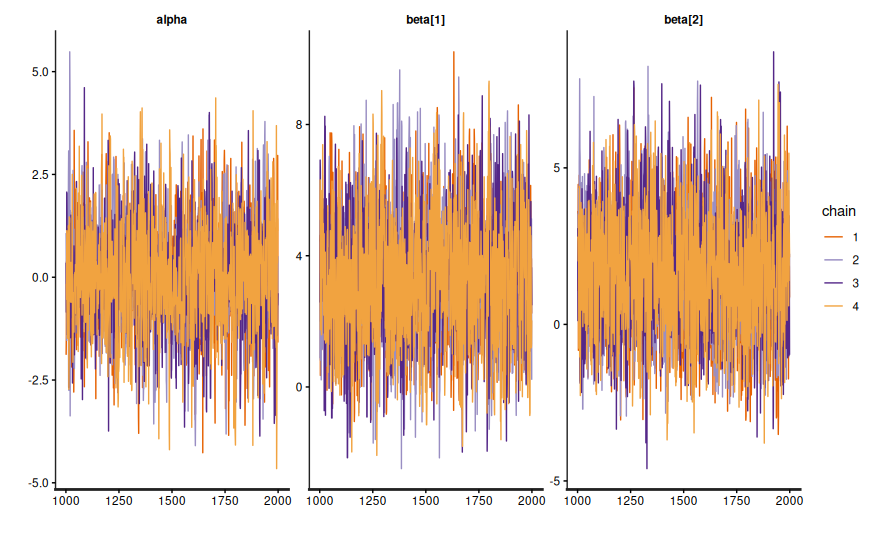
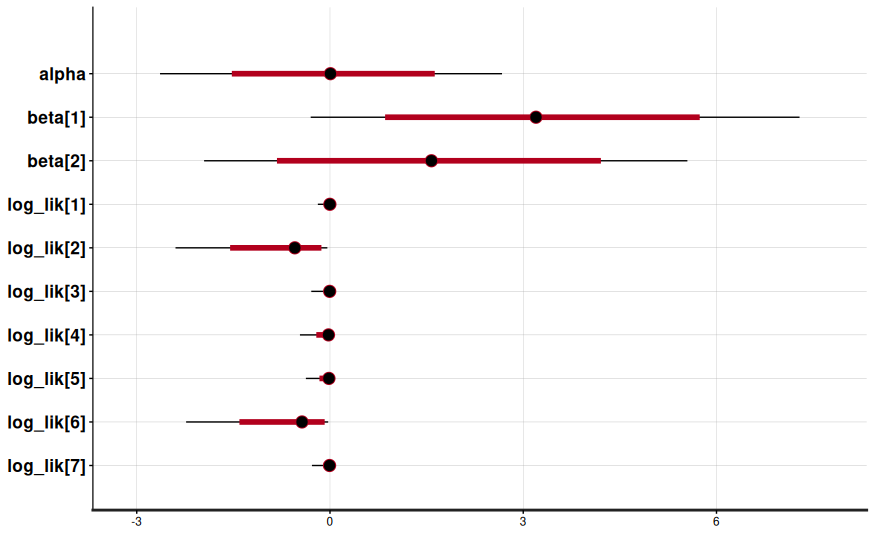
- Posterior predictive checks compare
y_hatdraws to the observed admissions. If the simulated acceptance rates miss the observed rate completely, revisit the model. - Leave-one-out cross-validation becomes trivial with the
log_likvector:arviz.loo(fit.to_inference_data())ranks competing specifications.
6. Extend the template
Logistic regression is just the opening move. Stan makes it straightforward to:
- Add hierarchical structure by letting intercepts or slopes vary by department (
alpha[dept] ~ normal(mu_alpha, sigma_alpha)). - Swap in regularizing priors (e.g.,
beta ~ normal(0, 1)or horseshoe priors) when predictors explode. - Incorporate interaction terms or nonlinear basis expansions before handing data to Stan.
- Move to ordered logistic or multinomial likelihoods when the response has more than two categories.
Each extension reuses the same data → model → sampling pipeline, so the initial investment pays off quickly.
7. Takeaways
- A small, fully worked example makes Stan approachable before tackling hierarchical or time-series models.
- Standardized predictors, explicit priors, and posterior predictive checks keep logistic regression fits stable.
- CmdStanPy bridges Python workflows with Stan’s compiled performance, while sharing Stan programs across teams.
- Diagnostics (
fit.diagnose(),R_hat, effective sample sizes) are first-class signals that the posterior is trustworthy. - The same pattern scales to richer discrete-outcome models—logistic regression is simply the friendliest starting point.